This tutorial will guide you through a complete image-based treatment simulation.
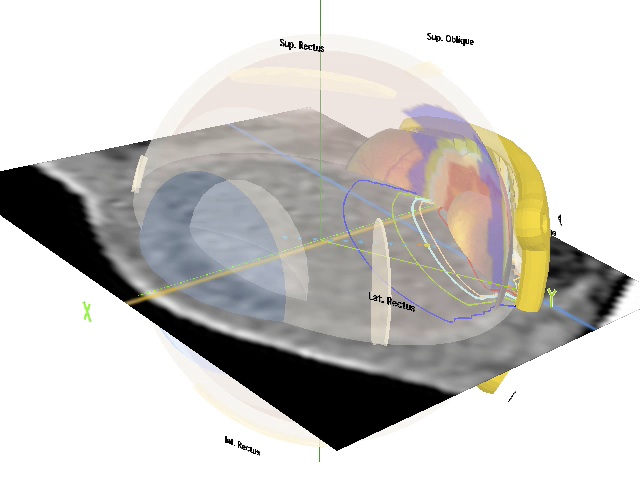
The case involves a rather small, posterior, choroidal melanoma located adjacent to the macula in a left eye. This case was selected for the tutorial because it utilizes and benefits from many of Plaque Simulator's technologies. You will gain experience:
- In the use of OsiriX to export multiplanar reconstructions (MPR) from 3D CT imaging to Plaque Simulator. Small ocular tumors are very difficult to image with CT or MR. In this case the tumor is visible in the CT images, albeit barely. The MPR images necessary for this tutorial are provided with Plaque Simulator.
- Fusing fundus photography with CT.
- Using ultrasound imaging.
- Fitting a notched plaque to the nearby optic nerve.
- Using a slot collimated plaque to spare the optic nerve.
- Conforming the source loading pattern to match the shape of the tumor base.
- Using the retina dose area histogram (RDAH) to evaluate dosimetric coverage of the tumor base and its surrounding margin.
- Adjusting prescription height to assure margin coverage.
- Calculating scleral suture coordinates.
- Printing documents.
You may choose whether to use a JavaScript movie controller, including a Play/Pause button and a clickable timeline, or the original QuickTime movie controller: