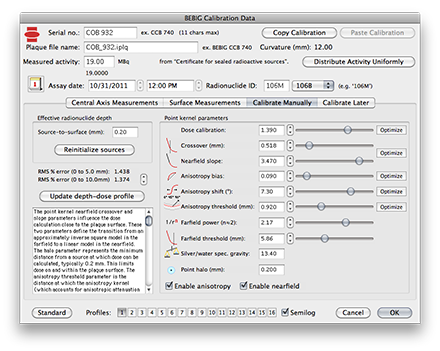
Manually Fine-Tuning the CalibratonAfter the automated optimization and surface annealing processes are complete you will need to manually fine tune the model parameters. Click the Calibrate Manually tab. Adjust the parameters using the sliders followed by the bumper arrows to minimize the RMS% error of the fit of the central axis profile to the measured data over the range 0..10mm. |
|

|

|
|
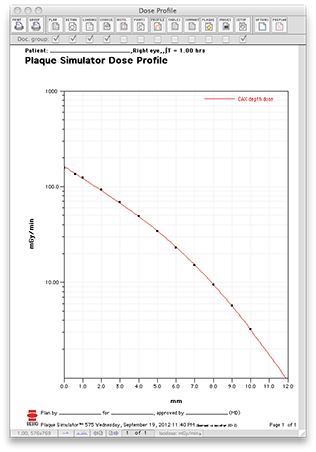
Below are sample isodose calculations for this plaque with its notch up against the optic nerve. All dosimetry has been normalized to 1.0 on the central axis of the plaque at 1 mm from the surface of the plaque, the same normalization point as the BEBIG near-surface measurements. Please note that the BEBIG points are normalized to a value of 100 whereas the PS isodose lines are normalized to 1.0. |